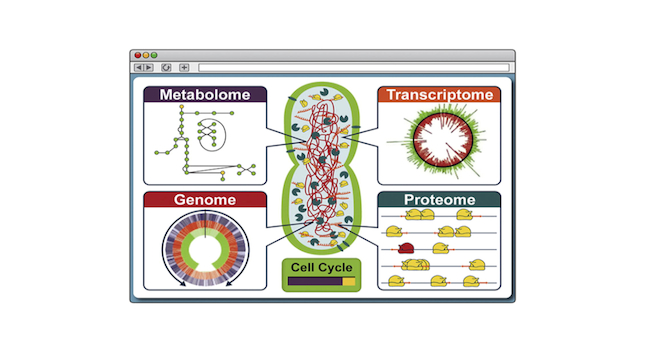
For the first time, researchers at Stanford University in California have used computer software to simulate the entire lifecycle of an organism — all 525 genes of the bacterium Mycoplasma genitalium.
The breakthrough, which is described in a paper published in the journal Cell on July 20, could lead to enormous advances in the fields of genetics, personalized medicine, pharmaceuticals and biology in general, allowing scientists to perform experiments that wouldn’t be possible on actual living organisms. Researchers used 128 computers to model an organism that is actually a sexually transmitted parasite.
“I’m a big believer in ‘model-driven discovery’ which simply means that if you have a model of a biological process, you will make discoveries more quickly and efficiently than you would without a model,” said Markus Covert, an assistant professor of bioengineering at Stanford, who led the study, in an email to TPM.
Specifically, Covert explained Stanford’s software simulation of the Mycoplasma genitalium lifecycle is distinct from previous efforts to model organisms using computers because of the vast volume of the data it encompasses — literally every single molecular process that takes place in the single-celled bacterium’s life.
Covert’s team has already begun experimenting with removing certain genes to see what happens in the simulation.
“Each simulation is one cell, one life cycle,” Covert said. “We’ve now run thousands of these, both for the normally growing cell as well as all of the single-gene knockout strains.”
The simulation takes about nine to 10 hours in order for one cell to divide, again emulating its real counterpart, as The New York Times reported about the study. The simulation stops when the organism “dies.”
“‘Death’ of the simulation is defined as an inability to divide, degradation of key components, etc,” Covert explained.
The Standford simulation of the parasite has already led to one important discovery: The finding that the longer it takes for a single cell to begin replicating its DNA in preparation for division, the shorter time it will actually take to replicate, which balances out to the same average time across all cells modeled.
Among the many new advances that the computer simulation could lead to is the possibility “for the wholesale creation of new microorganisms,” according to a Stanford news release on the work.
Still, it was a long road to get from the initial idea to simulate the bacterium’s life cycle on a computer to actually doing it, as covert explained.
“I first thought about getting involved in whole-cell modeling about thirteen years ago, when I read a quote by a scientist in the paper who said (paraphrasing) that the ultimate challenge in biology would be to create a computer model of a cell, because it would imply a fundamental understanding of how cells work,” Covert wrote. “I basically became obsessed with that quote, and still think about it every day!”
It took nearly four years to actually write the software to perform the simulation itself, and the use of over 900 prior scientific papers on Mycoplasma genitalium and other bacterium to create the perfect algorithm to simulate the organism. The research was funded in part by the National Institutes of Health.